//
//Softmax Regression.
//
//Syntax
softmax cat by x y z/
(options)
;
//Options
loop:nn //nn is number of loopmax for Newton Lapson method (default=1000)
eps:0.00xxx //0.00xx is congency condition of Newton improvemant value(default=0.000001)
//Example
get iris.csv@;


//Execute softmax instruct label of Species by iris data.
softmax Species by SepalL SepalW PetalL PetalW/
loop=500
;
//View Probability of target variable.
anaput ana1;
//Get average probability of each Species
get ana1;
statis prob_setosa prob_versicolor prob_virginica by Species;
anaput ana2;
get ana2;
if(_items_ == "AVERAGE") outrec;
put ana3;
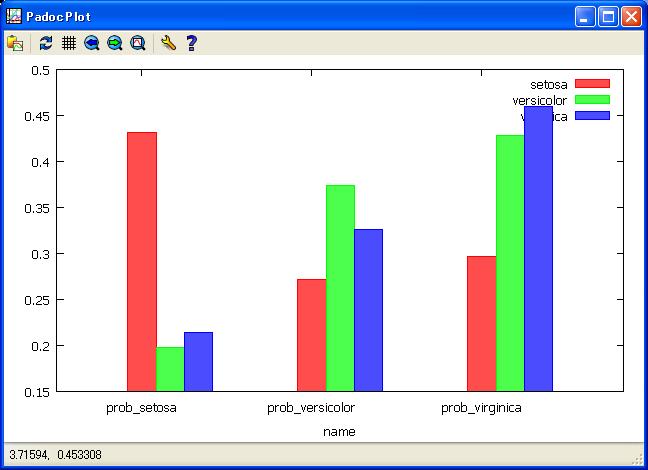
//View softmax average probablity by Species with few error classification
get ana3;
plot bar setosa versicolor virginica by name;
//view Weight for each cathcgorys
get socre@ana;
//Add nomal perturbation to iris data.
$set sig 1.0;
get iris.csv@;
sepall=SepalL*(1+ranstd(0,?sig));
petall=PetalL*(1+ranstd(0,?sig));
sepalw=SepalW*(1+ranstd(0,?sig));
petalw=PetalW*(1+ranstd(0,?sig));
put iris_sig@;
//Execute softmax normal perturbation iris data.
get iris_sig@;
softmax Species by sepall petall sepalw petalw/
eps=0.005
;
anaput ana5;
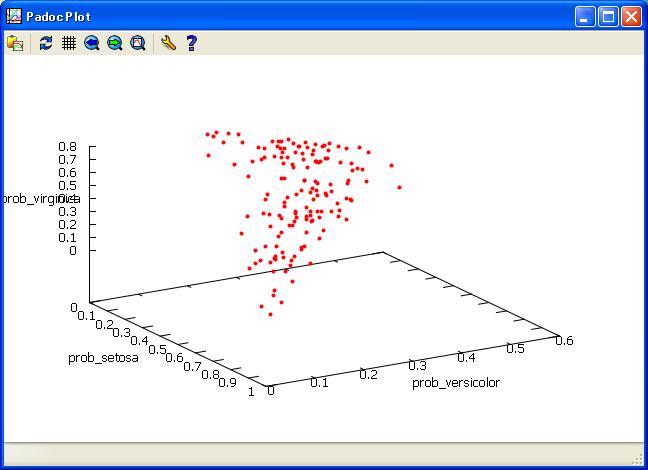
//view of Probability of Species.
plot scat prob_setosa prob_versicolor prob_virginica/
marker=OF
color=red
marksize=0.5
;
//Get average of probability of each Species.
get ana5;
statis prob_setosa prob_versicolor prob_virginica by Species;
anaput ana6;
get ana6;
if(_items_ == "AVERAGE") outrec;
put ana7;
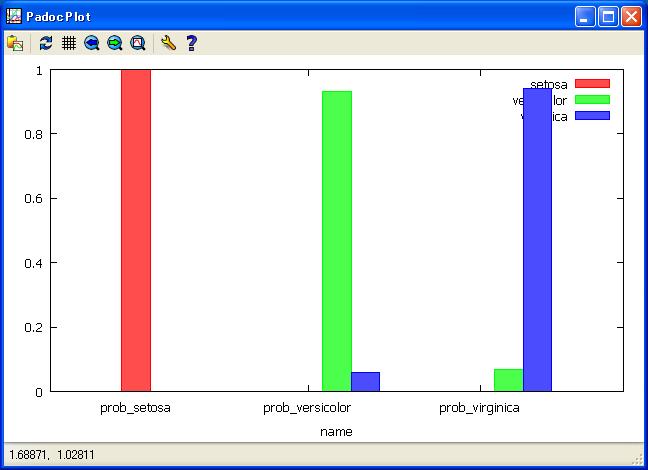
//Veiw softmax average probabilty by Species under normal perturbation.
get ana7;
plot bar setosa versicolor virginica by name;